Software
This is a list of software that can help you generate, consume or otherwise work with Bioschemas markup. If you have such software and you’d like to list it here, please email us at enquiries@bioschemas.org.
Note:
help us keep the above list updated: Join the community and/or create a pull request on the bioschemas github community project.
Bioschemas Generator
A web application for prototyping markup against the Bioschemas profiles.
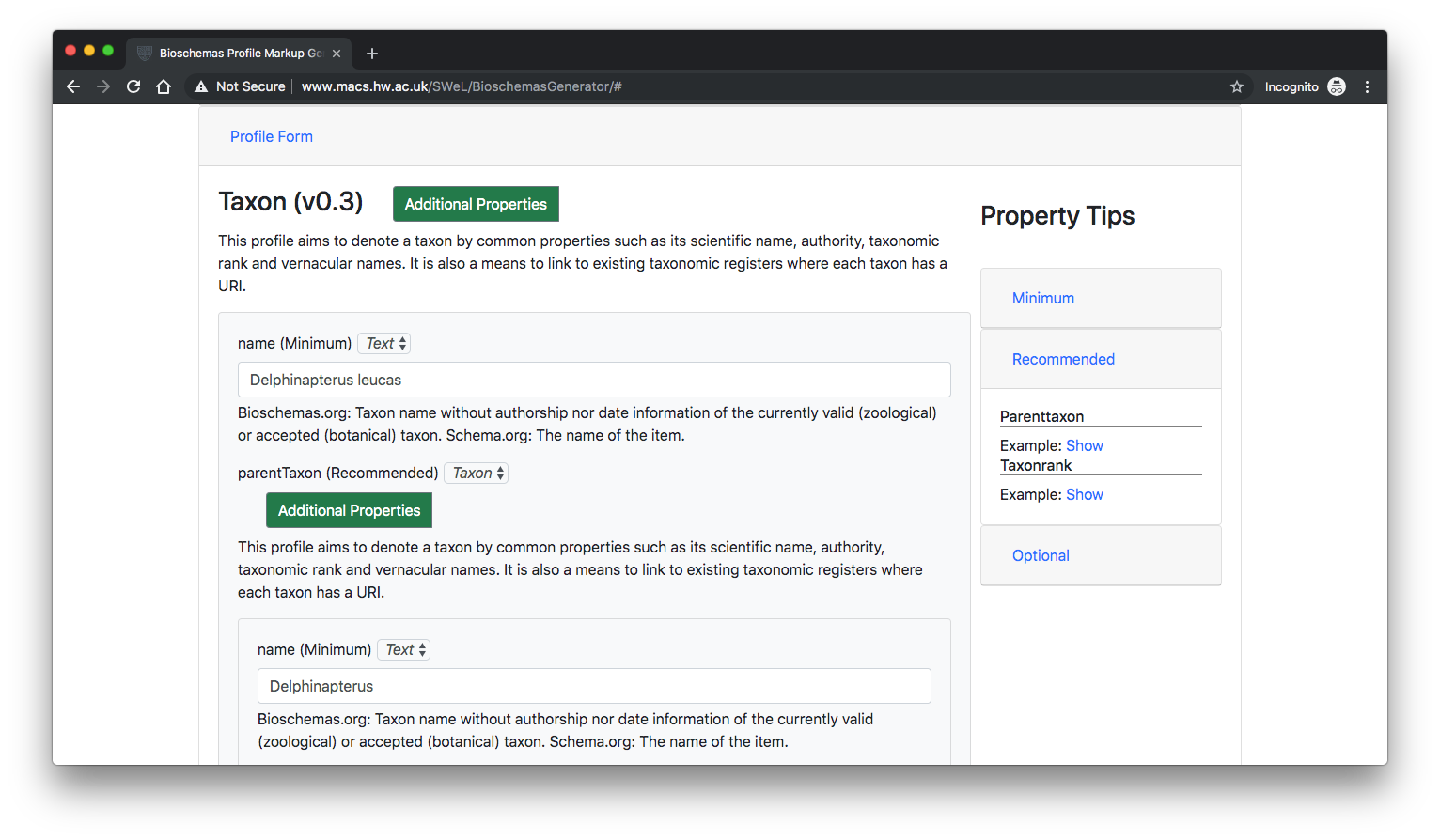
Description
This web application supports users in the creation of Bioschemas compliant markup required for inclusion on their web resource. Bioschemas provides profiles for Schema.org mark-up in order to structure and expose life-sciences metadata on the web. Each profile brings a list of allowed attributes with their constraints and properties. Some attributes are required, some are composite, some allow multiple values, some are under controlled vocabularies, and some can even be all of that.
The Bioschemas Generator is a web application that assists users in the creation of their metadata structure, through dynamically generated forms, allowing easier development of Bioschemas compliant markup for web resources.
People
Links
BMUSE: Bioschemas Markup Scraper and Extractor
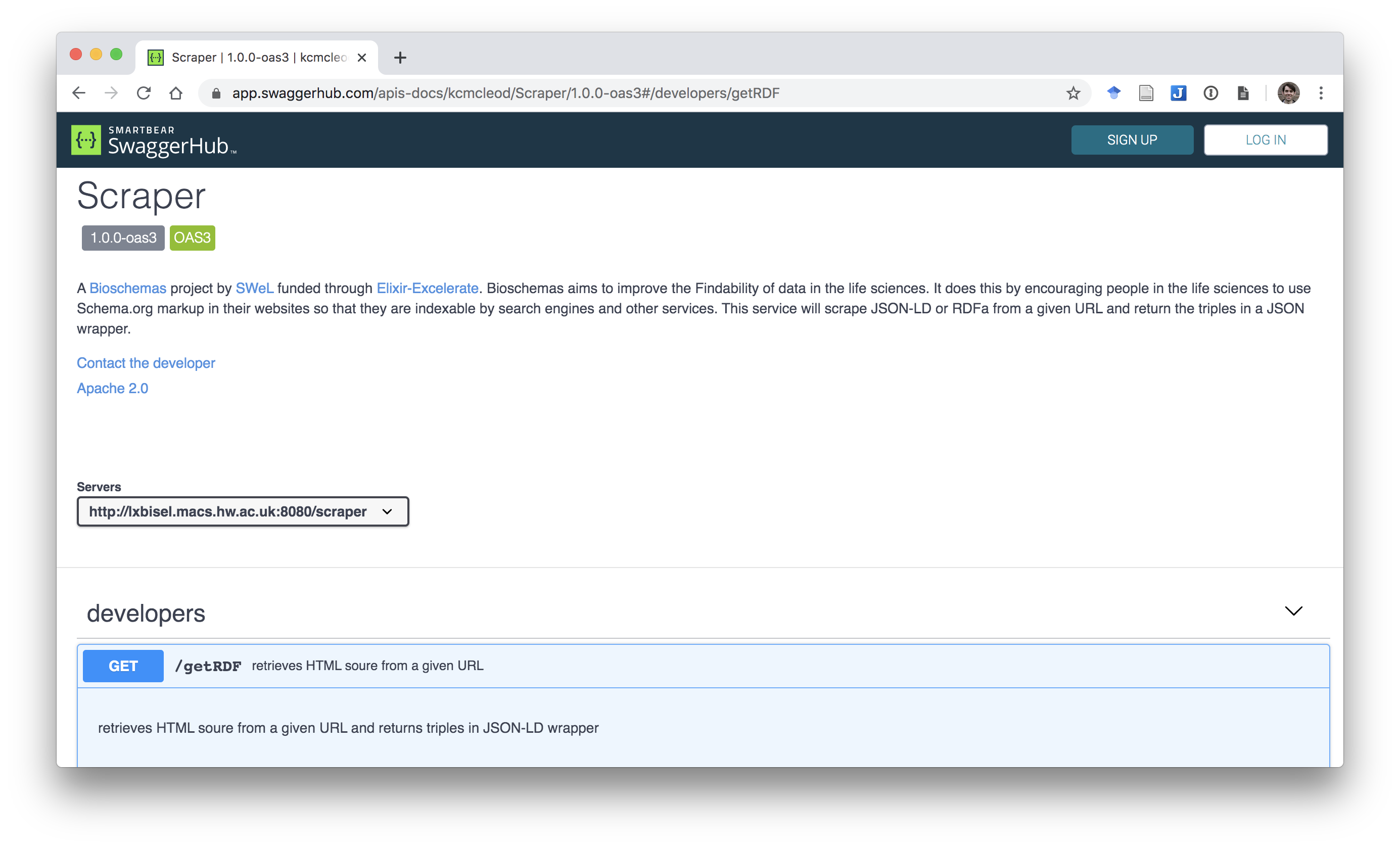
A scraper for extracting Bioschemas markup embedded in web sites.
Description
BMUSE is a directed scraping framework capable of extracting JSON-LD and RDFa markup from static and single page application sites. Markup is returned in a variety of RDF formats together with metadata about when the markup was extracted and from where.
BMUSE can be deployed as a scraping service. As input it takes a list of URLs or sitemaps. We have deployed a version to do a daily scrape of sites known to contain data relevant to COVID-19. You can get the data from here.
People
Links
- Try it out using our Swagger Hub deployment
- Get the code from GitHub
GoWeb
Application to help publishing Bioschemas profiles on the Bioschemas website.

Description
GoWeb is a simple application that helps us to create the Bioschemas website specifications. It extracts content from the google spreadsheets we use to define Bioschemas profiles (dataset, protein, samples, ...). The web specifications are created using YAML so they are also in a machine readable format. The combination of Google spreadsheets and GoWeb allows us to have a simple mechanism to define Bioschemas profiles and display them in a beautiful and consistent manner.
We have created a Google Spreadsheet template to help the community to define Bioschemas profiles. We have also described the process to create the web specifications using GoWeb.
People
Links
Google Dataset Search
Dataset specific search engine provided by Google.

Description
Bioschemas specifies certain Dataset properties as mandatory (name, description, keywords, etc.). These are a superset of the properties required by Google to index a dataset (name and description). Thus, a dataset marked up with Bioschemas mandatory properties, where that markup can be reached by a sitemap or site links, will also be available for Google's Dataset Search.
At Bioschemas we are working with Google and schema.org to improve existing schema.org types and add new ones that will improve the findability of life sciences data. This will enable search engines and other applications to provide relevant and comprehensive search results.
Links
Archive
A list of projects developed previously by the community that are now in hibernation.
GoCrawlIt
Minimal crawler and extractor of microdata and JSON-LD metadata.

Description
This tool provides a way to extract JSON-LD and microdata markup information from a given website.
People
Links
Validata
A web application for validating Bioschemas markup against the specifications.
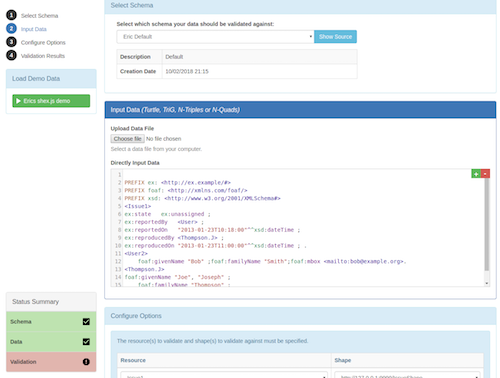
Description
This web app supports users in testing whether their generated markup is compliant with the Bioschemas specifications. The validation engine currently supports machine processable constraint descriptions written in the Shape Expression (ShEx) language which capture the Bioschema profiles. The Validata app allows users to supply some markup which is then compared to the profiles and generates an error report to the user for the snippet of code they provide.
People
- Alasdair Gray
- Leyla Garcia
- Nikita Filippov
Links
Buzzbang
Open-source components for software to find, crawl and use Bioschemas markup, and for humans to search it.

Description
The Buzzbang project develops components that other software can use to find, crawl and use Bioschemas markup. It also provides a Google-like frontend for humans to search it. It is at an early alpha stage but has already been used by some third-party prototype Bioschemas applications. Contributors and bug reports welcome!
People
Links
Markup Builder
A web application for prototyping markup against the Bioschemas profiles.
Description
This web application supports users in the creation of Bioschemas compliant markup required for inclusion on their web resource. Bioschemas provides profiles for schema.org mark-up in order to structure and expose life-sciences metadata on the web. Each profile brings a list of allowed attributes with their constraints and properties. Some attributes are required, some are composite, some allow multiple values, some are under controlled vocabularies and some can even be all of that. We want to create a web application that assist users into the creation of their metadata structure, through dynamically generated forms, allowing an easier sharing over the web.

